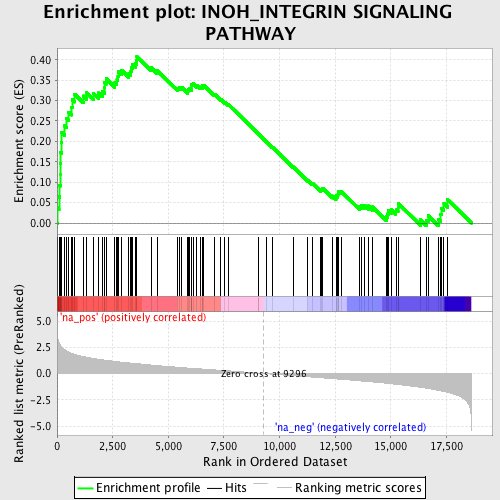
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_absentNotch_versus_wtNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | INOH_INTEGRIN SIGNALING PATHWAY |
| Enrichment Score (ES) | 0.40706855 |
| Normalized Enrichment Score (NES) | 1.8270376 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.6621815 |
| FWER p-Value | 0.63 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | PTK2 | 12 | 3.772 | 0.0381 | Yes | ||
| 2 | SDC4 | 86 | 2.957 | 0.0646 | Yes | ||
| 3 | RAF1 | 108 | 2.852 | 0.0928 | Yes | ||
| 4 | CD44 | 147 | 2.701 | 0.1185 | Yes | ||
| 5 | ITGB1 | 151 | 2.685 | 0.1459 | Yes | ||
| 6 | MAPK8 | 160 | 2.662 | 0.1729 | Yes | ||
| 7 | ITGB5 | 194 | 2.526 | 0.1971 | Yes | ||
| 8 | ITGA7 | 204 | 2.509 | 0.2224 | Yes | ||
| 9 | HSPG2 | 346 | 2.263 | 0.2380 | Yes | ||
| 10 | FYN | 416 | 2.163 | 0.2565 | Yes | ||
| 11 | DOCK10 | 520 | 2.048 | 0.2720 | Yes | ||
| 12 | MAPK10 | 659 | 1.913 | 0.2842 | Yes | ||
| 13 | LAMC2 | 686 | 1.892 | 0.3023 | Yes | ||
| 14 | COL18A1 | 794 | 1.821 | 0.3152 | Yes | ||
| 15 | DOCK8 | 1177 | 1.620 | 0.3113 | Yes | ||
| 16 | FN1 | 1326 | 1.556 | 0.3193 | Yes | ||
| 17 | COL3A1 | 1642 | 1.426 | 0.3169 | Yes | ||
| 18 | SDC3 | 1868 | 1.357 | 0.3187 | Yes | ||
| 19 | BCAN | 2033 | 1.305 | 0.3233 | Yes | ||
| 20 | VCL | 2133 | 1.276 | 0.3311 | Yes | ||
| 21 | ACAN | 2140 | 1.274 | 0.3438 | Yes | ||
| 22 | ITGAV | 2204 | 1.257 | 0.3534 | Yes | ||
| 23 | COL9A1 | 2599 | 1.158 | 0.3440 | Yes | ||
| 24 | RAP1B | 2690 | 1.135 | 0.3508 | Yes | ||
| 25 | COL1A2 | 2733 | 1.124 | 0.3601 | Yes | ||
| 26 | VCAN | 2753 | 1.117 | 0.3706 | Yes | ||
| 27 | RAP1A | 2888 | 1.081 | 0.3744 | Yes | ||
| 28 | MAP2K2 | 3227 | 1.012 | 0.3666 | Yes | ||
| 29 | SHC3 | 3316 | 0.993 | 0.3720 | Yes | ||
| 30 | COL4A1 | 3354 | 0.981 | 0.3801 | Yes | ||
| 31 | COL8A2 | 3379 | 0.976 | 0.3889 | Yes | ||
| 32 | NRAS | 3511 | 0.954 | 0.3916 | Yes | ||
| 33 | SMC3 | 3572 | 0.942 | 0.3981 | Yes | ||
| 34 | GRB2 | 3585 | 0.939 | 0.4071 | Yes | ||
| 35 | CRK | 4221 | 0.808 | 0.3811 | No | ||
| 36 | KRAS | 4500 | 0.757 | 0.3739 | No | ||
| 37 | COL2A1 | 5414 | 0.594 | 0.3307 | No | ||
| 38 | MAPK9 | 5502 | 0.579 | 0.3320 | No | ||
| 39 | ITGA5 | 5612 | 0.565 | 0.3319 | No | ||
| 40 | DOCK7 | 5876 | 0.520 | 0.3230 | No | ||
| 41 | SDC1 | 5886 | 0.519 | 0.3279 | No | ||
| 42 | ITGA11 | 5966 | 0.506 | 0.3288 | No | ||
| 43 | RAC1 | 6030 | 0.496 | 0.3305 | No | ||
| 44 | ITGB6 | 6031 | 0.496 | 0.3356 | No | ||
| 45 | DOCK9 | 6035 | 0.495 | 0.3405 | No | ||
| 46 | COL8A1 | 6110 | 0.484 | 0.3415 | No | ||
| 47 | ITGAX | 6278 | 0.457 | 0.3372 | No | ||
| 48 | ITGA6 | 6435 | 0.438 | 0.3333 | No | ||
| 49 | COL4A2 | 6520 | 0.426 | 0.3331 | No | ||
| 50 | ELN | 6535 | 0.422 | 0.3367 | No | ||
| 51 | COL1A1 | 6590 | 0.413 | 0.3381 | No | ||
| 52 | COL10A1 | 7070 | 0.343 | 0.3157 | No | ||
| 53 | BGN | 7355 | 0.306 | 0.3035 | No | ||
| 54 | BCAR1 | 7545 | 0.278 | 0.2962 | No | ||
| 55 | DOCK2 | 7706 | 0.254 | 0.2902 | No | ||
| 56 | MAP2K1 | 9029 | 0.041 | 0.2192 | No | ||
| 57 | CAV1 | 9067 | 0.034 | 0.2176 | No | ||
| 58 | DOCK4 | 9394 | -0.016 | 0.2002 | No | ||
| 59 | ITGA2B | 9696 | -0.059 | 0.1845 | No | ||
| 60 | SDC2 | 10605 | -0.203 | 0.1376 | No | ||
| 61 | LUM | 11257 | -0.299 | 0.1055 | No | ||
| 62 | COL6A1 | 11484 | -0.337 | 0.0968 | No | ||
| 63 | ITGB3 | 11860 | -0.399 | 0.0806 | No | ||
| 64 | SRC | 11895 | -0.404 | 0.0830 | No | ||
| 65 | ITGA3 | 11933 | -0.410 | 0.0852 | No | ||
| 66 | COL6A2 | 12375 | -0.483 | 0.0663 | No | ||
| 67 | COL12A1 | 12537 | -0.506 | 0.0628 | No | ||
| 68 | COL11A1 | 12547 | -0.509 | 0.0676 | No | ||
| 69 | COL11A2 | 12622 | -0.524 | 0.0690 | No | ||
| 70 | GSG2 | 12628 | -0.524 | 0.0741 | No | ||
| 71 | KERA | 12665 | -0.530 | 0.0776 | No | ||
| 72 | LCK | 12782 | -0.552 | 0.0770 | No | ||
| 73 | SHC1 | 13608 | -0.694 | 0.0396 | No | ||
| 74 | ITGAM | 13674 | -0.706 | 0.0434 | No | ||
| 75 | ITGA4 | 13801 | -0.729 | 0.0441 | No | ||
| 76 | DCN | 13985 | -0.760 | 0.0420 | No | ||
| 77 | ITGAE | 14182 | -0.800 | 0.0397 | No | ||
| 78 | LAMB3 | 14798 | -0.920 | 0.0159 | No | ||
| 79 | TLN1 | 14857 | -0.932 | 0.0224 | No | ||
| 80 | VAV1 | 14880 | -0.936 | 0.0308 | No | ||
| 81 | ITGB7 | 15011 | -0.966 | 0.0337 | No | ||
| 82 | MAPK3 | 15232 | -1.017 | 0.0323 | No | ||
| 83 | ITGAL | 15325 | -1.039 | 0.0380 | No | ||
| 84 | GPC4 | 15330 | -1.040 | 0.0485 | No | ||
| 85 | RHOA | 16330 | -1.305 | 0.0080 | No | ||
| 86 | DOCK3 | 16606 | -1.393 | 0.0074 | No | ||
| 87 | NCAN | 16680 | -1.421 | 0.0181 | No | ||
| 88 | CDC42 | 17148 | -1.605 | 0.0094 | No | ||
| 89 | HRAS | 17243 | -1.641 | 0.0212 | No | ||
| 90 | SOS2 | 17269 | -1.650 | 0.0368 | No | ||
| 91 | SOS1 | 17388 | -1.701 | 0.0479 | No | ||
| 92 | ITGB2 | 17553 | -1.780 | 0.0574 | No |